Research Summary
Defining the mechanisms of virus replication and understanding the role of immune cells and host factors in regulating viral infection are essential for developing effective therapeutic strategies. Dr. Wu’s lab studies the mechanisms of HIV and SARS-CoV-2 replication, virus interaction with host factors, and anti-viral innate immunity. The lab focuses on the mechanisms of HIV-1 restriction by the host protein SAMHD1, a unique cellular dNTP triphosphohydrolase that regulates dNTP homeostasis and maintains genomic stability. The lab also investigates the mechanisms by which N6-methyladenosine (m6A) modifications of viral and cellular RNA regulate virus gene expression and the host cell response to infection. These mechanistic studies can lay the foundation to facilitate the development of more effective interventions against viral infections.
Our current projects are:
1) SAMHD1-mediated regulation of HIV-1 innate immunity and viral gene expression. Our central hypothesis is that SAMHD1 modulates HIV-1 antiviral immunity and viral gene expression by suppressing NF-kB activation, type-I interferon induction, and viral transcription. Studying how SAMHD1 regulates HIV-1 infection, innate immune responses, and viral gene expression will provide new insights into the development of more effective strategies to eradicate HIV-1 infection.
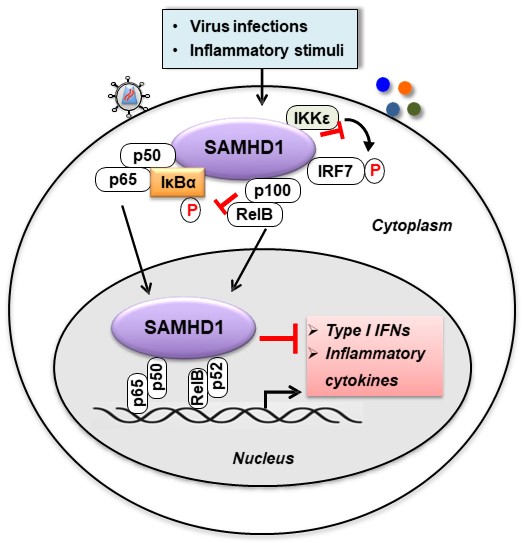
Chen, S., et al. (2018). SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-kB and interferon pathways. PNAS, 115(16), E3798-E3807. PMID: 29610295.
SAMHD1 enhances HIV-1-induced apoptosis in monocytic cells via the mitochondrial pathway. mBio.2025 May 28;:e0042525. doi: 10.1128/mbio.00425-25. PMID: 40434097.
dNTP depletion and beyond: the multifaceted nature of SAMHD1-mediated viral restriction. J Virol.2025 May 20;99(5):e0030225. doi: 10.1128/jvi.00302-25. Epub 2025 Apr 25. Review. PubMed PMID: 40277359.
2) Targeting HIV-1 RNA modifications in latently infected CD4+ T cells for therapeutic development. Despite highly effective anti-retroviral therapy (ART), the latent HIV-1 reservoir in resting CD4+ T cells is the major barrier to a functional cure of HIV-1 infection. This project aims to understand the mechanisms of post-transcriptional regulation of HIV-1 RNA in HIV-1-infected individuals who are on ART, and to develop a novel therapeutic strategy to alter post-transcriptional RNA modifications as a potential therapeutic platform for inhibiting HIV-1 replication.
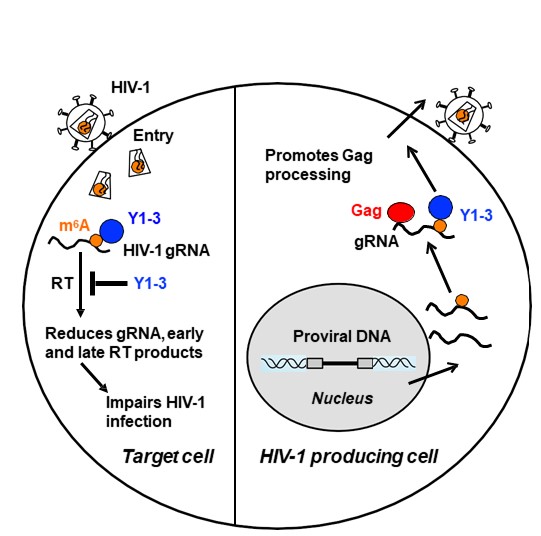
Lu, W., et al. (2018). N6-Methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. J Biol Chem, 293(34), 12992-13005. PMID: 29976753.
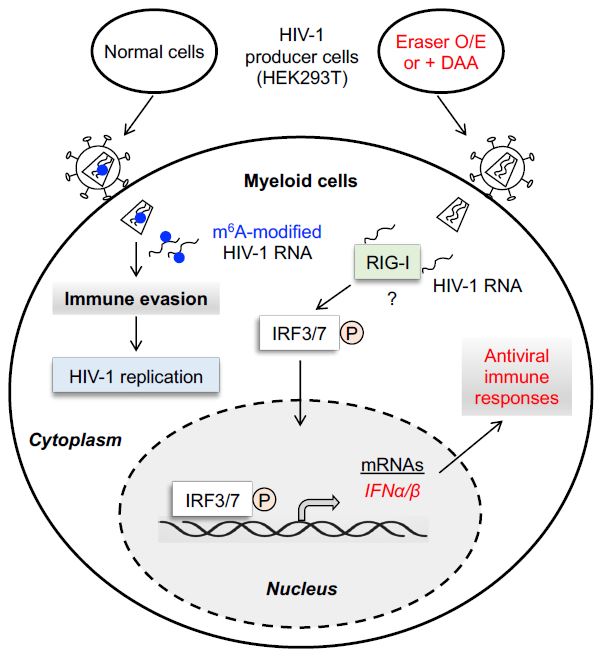
Chen S., et al. (2021). N6-methyladenosine modification of HIV-1 RNA suppresses type-I interferon induction in differentiated monocytic cells and primary macrophages. PLoS Pathogens, 17(3):e1009421. PMID: 33690734
Epitranscriptomic m(6)A modifications during reactivation of HIV-1 latency in CD4(+) T cells. mBio. 2024;15(11):e0221424. doi: 10.1128/mbio.02214-24. PMID: 39373537
Single-molecule epitranscriptomic analysis of full-length HIV-1 RNAs reveals functional roles of site-specific m(6)As. Nat Microbiol. 2024;9(5):1340-1355. doi: 10.1038/s41564-024-01638-5. PubMed PMID: 38605174
3) HIV-1-induced upregulation of m6A modifications of cellular RNA in CD4+ T-cells. To investigate the function and mechanisms of HIV-1-induced upregulation of m6A modifications of cellular RNA in primary CD4+ T cells. Defining m6A epitranscriptomic and general transcriptomic profiles of CD4+ T cells will reveal cellular RNAs involved in regulating HIV-1 infection.
4) SAMHD1-mediated Regulation of Innate Immunity During SARS-CoV-2 Infection. To examine the role of SAMHD1 in SARS-CoV-2-induced inflammation and innate immunity.